Sep 28 2020
With a new optical switch, scientists can now accurately control the lifespan of genetic “copies.” Such genetic “copies” are used by the cell as building instructions for protein synthesis.
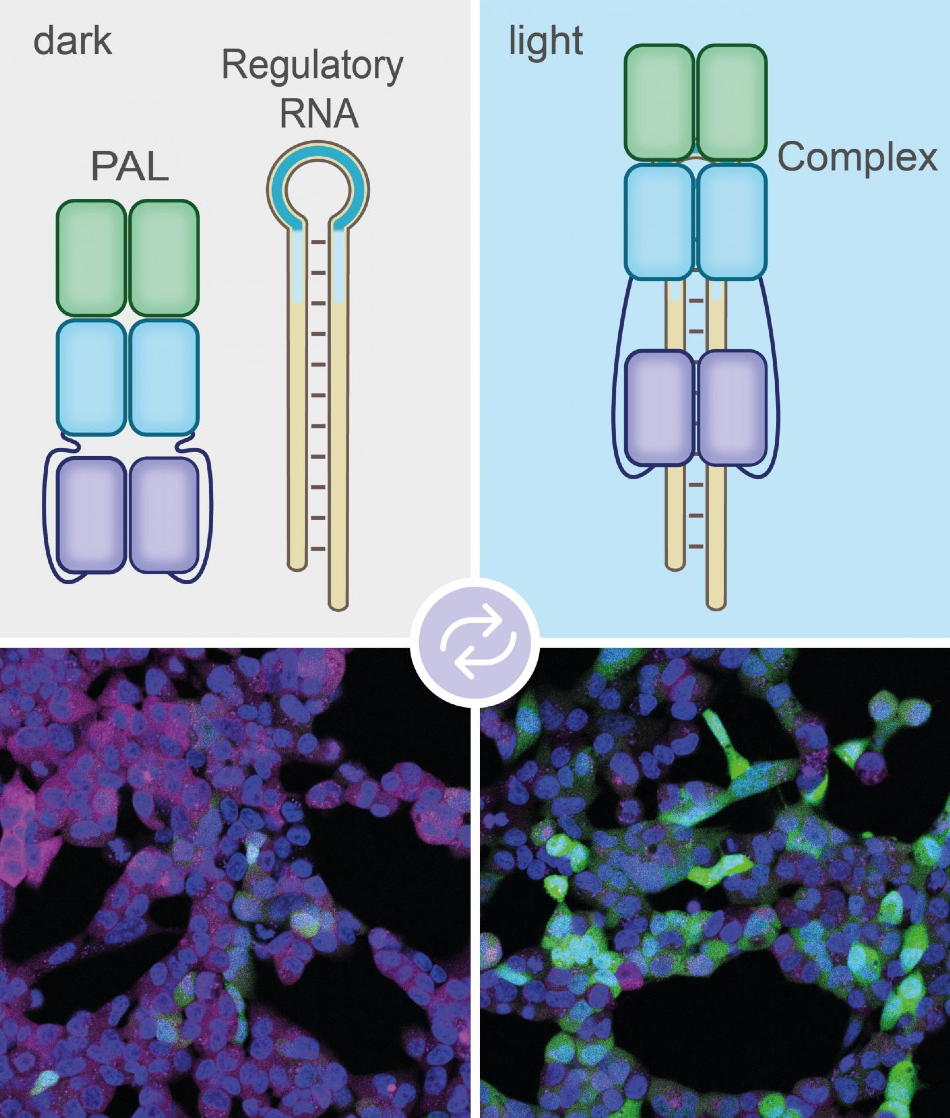 When illuminated the PAL molecule binds to the aptamer (blue loop at the top left). The label made of regulatory RNA can therefore no longer bind to the mRNA. This way it will not be degraded. Image Credit: © Sebastian Pilsl/AG Mayer/University of Bonn.
When illuminated the PAL molecule binds to the aptamer (blue loop at the top left). The label made of regulatory RNA can therefore no longer bind to the mRNA. This way it will not be degraded. Image Credit: © Sebastian Pilsl/AG Mayer/University of Bonn.
The technique was designed by scientists from the University of Bonn and the University of Bayreuth. It may considerably advance the analysis of dynamic processes that occur in living cells. The new study has been published in the Nature Communications journal.
Metaphorically speaking, a huge library of an unlimited number of books, called genes, is present in the nucleus of each human cell. Each of these books, consequently, contains the building instructions for a specific protein. When a specific protein is required by the cell, a transcription of the equivalent instructions is made. Such transcriptions are known as mRNAs (RNA is a slightly altered form of DNA).
A cellular mechanism makes sure that the mRNA transcriptions are “shredded” again after a brief period. This process ensures that the protein is only created as long as it is really required. Many years ago, scientists developed a concept of applying this shredder for their purposes—by specifically binding a marker to specific mRNAs, they make sure that the transcriptions are never utilized as building instructions but are damaged instantly—a process also called RNA silencing. The cell subsequently lacks the equivalent protein. This renders it possible to determine the kind of function it would actually be accountable for.
Bacterial Molecule as Light-Dependent Switch
The method, recently reported by the research team from the universities of Bonn and Bayreuth, is based on this technique. While it is nowhere near as crude, it enables a highly differentiated control across the lifespan of the mRNA copies.
We use a bacterial molecule to control the shredding of mRNA transcriptions with the help of light.
Dr Günter Mayer, Professor and Head, Chemical Biology & Medicinal Chemistry Research Group, LIMES Institute, University of Bonn
The bacterial molecule with the abbreviation PAL serves as a type of switch. It alters its shape under the effect of blue light. During the process, a pocket is exposed that can attach to specific molecules.
We searched a huge library of artificially produced short RNA molecules called aptamers. Eventually we came across an aptamer that's a good match for the pocket in the PAL molecule.
Dr Günter Mayer, Professor and Head, Chemical Biology & Medicinal Chemistry Research Group, LIMES Institute, University of Bonn
Currently, the team has combined this aptamer with one of the molecular markers that is capable of binding to mRNAs and thus discharges them for degradation.
When we irradiate the cell with blue light, PAL binds to the marker via the aptamer and thus puts it out of action. The mRNA is then not destroyed, but translated into the corresponding protein.
Sebastian Pilsl, University of Bonn
Pilsl is also a colleague of Mayer.
Once the researchers turned off the blue light, PAL discharges the label again. It can now adhere itself to the mRNA, which is further shredded.
In the future, this will allow scientists to precisely examine when and where a protein is required in a cell, by merely immersing a specific cell area in blue light at a certain time and then observing the consequences.
In the present study, the researchers applied this to proteins that play a crucial role in the regulation of the cell division and cell cycle. Through genetic engineering, the combination of degradation marker and aptamer is introduced into the cell. This implies that it produces the light-dependent degradation signal by itself, and does not have to be delivered from outside.
Gene Transcriptions can be Specifically Switched off
The aptamer can be integrated with any markers, each of which consequently acts as a shredder signal for a particular mRNA.
“This method can therefore be used to switch off practically every mRNA molecule in the cell in a controlled manner,” emphasized Professor Dr Andreas Möglich from the University of Bayreuth.
In the newly published pilot study, it all worked both easily and reliably. The team thus sees an excellent potential in their technique for the analysis of dynamic processes in living organisms and cells.
Journal Reference:
Pilsl, S., et al. (2020) Optoribogenetic control of regulatory RNA molecules. Nature Communications. doi.org/10.1038/s41467-020-18673-5.