Nov 3 2012
The ability of fluorescence microscopy to study labeled structures like cells has now been empowered to deliver greater spatial and temporal resolutions that were not possible before, thanks to a new method developed by University of Illinois researcher Gabriel Popescu and Ru Wang from his lab. Using this method, they were able to study the critical process of cell transport dynamics at multiple spatial and temporal scales and reveal, for the first time, properties of diffusive and directed motion transport in living cells.
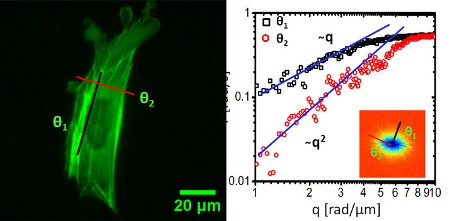 Dispersion-relation fluorescence spectroscopy of mouse embryonic fibroblast (MEF): a) fluorescence image showing a cell whose actin was labeled with GFP. b) Dispersion curve measured for the cell in a. The black and red lines indicate directed motion (along the actin filaments) and diffusion (perpendicular to the actin filaments). Inset shows the dispersion map. (Image courtesy Gabriel Popescu)
Dispersion-relation fluorescence spectroscopy of mouse embryonic fibroblast (MEF): a) fluorescence image showing a cell whose actin was labeled with GFP. b) Dispersion curve measured for the cell in a. The black and red lines indicate directed motion (along the actin filaments) and diffusion (perpendicular to the actin filaments). Inset shows the dispersion map. (Image courtesy Gabriel Popescu)
Popescu leads the Quantitative Light Imaging Laboratory at Illinois' Beckman Institute, while Wang of the lab is first author on the paper reporting the method in Physical Review Letters. The new approach, called dispersion-relation fluorescence spectroscopy (DFS), labels molecules of interest with a fluorophore whose motion, the researchers write, "gives rise to spontaneous fluorescence intensity fluctuations that are analyzed to quantify the governing mass transport dynamics. These data are characterized by the effective dispersion relation."
That ability to study the directed and diffusive transport characteristics of cellular dispersion through a wide range of temporal and spatial scales is more comprehensive than using just fluorescence microscopy. It provides more information than existing methods, such as fluorescence correlation spectroscopy (FCS), which is widely used for studying molecular transport and diffusion coefficients at a fixed spatial scale.
This study used DFS to focus on the cell cytoskeleton subunit actin and found that "the fluorescently labeled actin cytoskeleton exhibits active transport motion along a direction parallel to the fibers and diffusive on the perpendicular direction." Those results, the researchers said, describe at what scale and when directed versus diffusive motion is taking place in the cell.
"So for the first time we think we're able to tell those apart and the spatial scales at which each is dominant," Popescu said.
"Some traditional methods are good at measuring local transport and some are good at measuring the larger scales," Wang said. "Our method gives a fuller view of what happens inside the cell, to the patterns of traffic. So we can look at both the local scale and at larger scales, and ask at which scale the motion transitions from random to directed motion."
Popescu said the multiplicity of scales the method offers over techniques like fluorescence correlated spectroscopy is key. Such knowledge would be valuable for researchers interested in the basic science of cellular dynamics, as well as those working in biomedical research, such as in analysis of a drug's effect on the body. This technique can be used with current fluorescence microscopy methods.
"I think that the beauty of this method is that you can use a commercial fluorescent microscope that is found everywhere to collect and analyze data in a very simple way," Wang said. "You don't need complicated expertise. Everyone can use it."
The method relies on taking time-resolved sequential data from fluorescent spectroscopic microscopy images and transforming them using the Fourier transform. This computational method enables easier understanding of the image data, providing a different representation of the image. Taking advantage of the respective frequency domains of patterns in the data, as this method does, is especially useful for trying to understand cellular dynamics like transport.
"So for the first time we saw this universal transport behavior in a living system: a clear combination of diffusive transport, like Brownian motion, and directed, deterministic transport," Popescu said. "As a general trend, we found that diffusion is dominant at short scales and directed transport at large distances."