Oct 22 2012
The glowing green molecule known as green fluorescent protein (GFP) has revolutionized molecular biology. When GFP is attached to a particular protein inside a cell, scientists can easily identify and locate it using fluorescence microscopy. However, GFP can’t be used with electron microscopy, which offers much higher resolution than fluorescence microscopy.
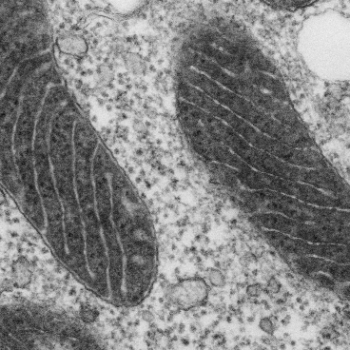 In this electron microscope image of a mitochondrion, the matrix has been stained with APEX, making it appear dark. The lighter projections into the matrix represent the intermembrane space. Image: Tom Deerinck and Jeff Martell
In this electron microscope image of a mitochondrion, the matrix has been stained with APEX, making it appear dark. The lighter projections into the matrix represent the intermembrane space. Image: Tom Deerinck and Jeff Martell
Chemists from MIT have now designed a GFP equivalent for electron microscopy — a tag that allows scientists to label and visualize proteins with unprecedented clarity.
“With things that may appear only a few pixels across by fluorescence microscopy — for example, a mitochondrion — you can’t make out any of the internal features. But with electron microscopy it’s very easy to discern the intricate internal structures,” says Jeff Martell, a graduate student in chemistry at MIT and lead author of a paper describing the new tag in the Oct. 21 online edition of Nature Biotechnology.
The new tag could help scientists pinpoint the locations of many cell proteins, providing new insight into those proteins’ functions, according to the researchers.
Improving on nature
Dubbed APEX, the new tag is similar to naturally occurring proteins that have been tried as imaging labels for electron microscopy. Horseradish peroxidase (HRP) is one commonly used tag, but it works only in a few compartments of a cell. Other recently developed tags work throughout a cell but are technically challenging to use because they require light to be shined on the sample and oxygen to be bubbled through it.
To improve on these methods, the researchers started with a protein similar to HRP, called ascorbate peroxidase (APX). APX is more versatile than HRP because it can function within a cell’s cytosol, in the main cavity of a cell.
Both HRP and APX belong to a class of enzymes called peroxidases, which remove an electron and a proton from other molecules in a process known as oxidation. Every peroxidase has different targets, and one of HRP’s main targets is a molecule called DAB, which when oxidized can be visualized with electron microscopy. The researchers genetically engineered APX so that it would also target DAB.
To use this new APEX tag (for “engineered APX”), the researchers deliver, into a living cell, a small ring of DNA containing the APEX gene joined to the gene for the protein they plan to image. The cell then produces the target protein, bound to the APEX protein.
Next, the researchers need to deliver DAB, which is not normally found in cells. This delivery takes place during the process of “fixing,” or stabilizing cells, which must be done before they can be imaged with electron microscopy.
When the APEX protein oxidizes DAB, it generates radicals that rapidly clump together into a tarlike polymer. That polymer can be detected through electron microscopy, allowing the researchers to pinpoint the location of the target protein.
A biological question resolved
To demonstrate the usefulness of their new tag, the researchers set out to resolve an open question regarding the location of a calcium channel protein discovered last year. Two research groups identified the protein and reported that it is located within mitochondria, but they had conflicting theories as to its precise location and orientation. Using the new imaging technique, the MIT-led team labeled the protein and determined that it is embedded in the inner mitochondrial membrane and faces into the innermost part of mitochondria, the mitochondrial matrix.
The team also showed that the new tag can label proteins throughout the cell — not only within mitochondria but also in the nucleus, the endoplasmic reticulum and the cytosol.
Martell and Alice Ting, the Ellen Swallow Richards Associate Professor of Chemistry at MIT and senior author of the Nature Biotechnology paper, invented the new technology. Other authors who helped to test the tag and explore biological applications are Mark Ellisman, Thomas Deerinck and Gina Sosinsky of the University of California at San Diego, Yasemin Sancak and Vamsi Mootha of Harvard Medical School, and Thomas Poulos of the University of California at Irvine.
In current studies, the researchers are working on filling entire cells, such as neurons, with their imaging agent. This allows certain neurons in an electron microscope image to stand out, making it easier to trace the connections they make with other neurons. For that project, the MIT researchers are collaborating with Joshua Sanes, a professor of molecular and cellular biology at Harvard University, who says he believes the new labeling technology will be very useful.
“We want to find the exact connections that these cells are making, and APEX is a good way to label cells for electron microscopy. We can label specific types of cells and figure out how they fit into the neural circuitry,” Sanes says.
Ting and Martell have filed for a patent on their imaging technology and are now working on making the APEX molecule more stable and better able to bind heme (an iron atom embedded in an organic compound), which is necessary for it to function properly.