Mar 10 2020
A researcher from Clemson University’s College of Science, in association with a research team mainly from Germany-based Heinrich Heine University, has created and shown an innovative optical imaging technique to track a solitary molecule in action.
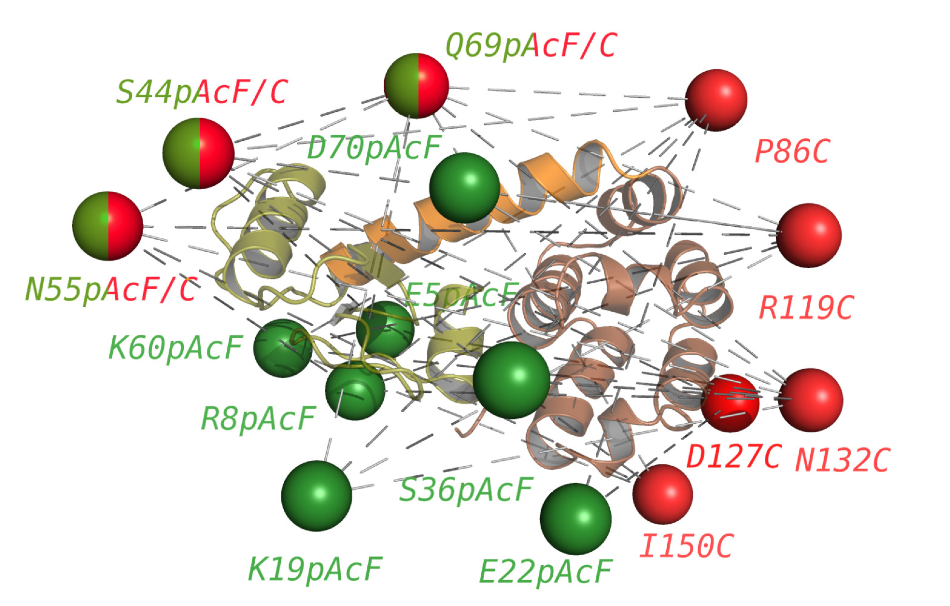 The researchers engineered 34 different double-labeled samples for FRET experiments to measure distances across the enzyme to visualize its function. Each sphere represents one of the fluorescence markers with the corresponding engineered mutation site. Image Credit: Hugo Sanabria.
The researchers engineered 34 different double-labeled samples for FRET experiments to measure distances across the enzyme to visualize its function. Each sphere represents one of the fluorescence markers with the corresponding engineered mutation site. Image Credit: Hugo Sanabria.
The fluorescence-based method may speed up the area of structural biology and help researchers to get a deeper understanding of the way molecules function, communicate, and assemble, which, consequently, may help in designing structure-guided drugs.
Together with his collaborators, Hugo Sanabria, an associate professor of physics and astronomy at Clemson University, utilized Förster resonance energy transfer (FRET) to analyze the lysozyme of the bacteriophage T4.
The researchers reported their findings in the study titled “Resolving dynamics and function of transient states in single enzyme molecules,” published in the Nature Communications journal on March 6th, 2020.
Claus A.M. Seidel, the study’s co-author and chair of the Institute for Molecular Physical Chemistry at Heinrich Heine University based in Germany, stated that this study underpins the vital reaction steps of biomolecular machines (or enzymes).
Our FRET studies demonstrate the need of a third functional state in the famous Michaelis–Menten kinetics. The Michaelis–Menten description is one of the best-known models of enzyme kinetics.
Claus A.M. Seidel, Study Co-Author and Chair, Institute for Molecular Physical Chemistry, Heinrich Heine University
A FRET-based microscope forms the heart of this imaging tool. It is a powerful and advanced instrument that can visualize biomolecules down to a few nanometers.
To observe biomolecules at work, Sanabria and collaborators positioned a pair of fluorescent markers on a group of molecules, which produced a ruler at the molecular level. The researchers used different sites of the markers and eventually gathered a series of distances that elucidate the form and shape of the visualized molecule.
This procedure essentially produced a set of data points that were computationally processed and enabled the scientists to differentiate how the molecule moves and how it looks.
We observe changes in the structure, and because our signal is time-dependent, we can also get an idea of how the molecule is moving over time.
Hugo Sanabria, Associate Professor, Department of Physics and Astronomy, Clemson University
In this latest research, Sanabria and his group of collaborators integrated the FRET-based microscope along with molecular simulations to analyze lysozyme—an enzyme present in mucus and tears. This enzyme damages the protective chains of carbohydrates surrounding the cell wall of bacteria.
Lysozyme is extensively used by researchers to explore the function and structure of proteins because it happens to be a stable enzyme.
“We can track the lysozyme of the bacteriophage T4 as it processes its substrate at near atomistic level with unprecedented spatial and temporal resolution,” added Sanabria. “We’ve taken the imaging field to a whole new level.”
The optical technique developed by Sanabria showed that the structure of lysozyme is different than contemplated earlier. To date, researchers have mostly established the structures of proteins, such as lysozyme, by primarily using techniques like cryo-electron microscopy, nuclear magnetic resonance (NMR) spectroscopy, and X-ray crystallography.
For the longest time, this molecule was considered a two-state molecule because of how it receives the substrate or cell wall of the target bacteria. However, we have identified a new functional state.
Hugo Sanabria, Associate Professor, Department of Physics and Astronomy, Clemson University
A database is now being established by the researchers, which would be used to store the team’s FRET-based structural models of analogously produced biomolecular models. These models could also be accessed by other investigators. Along with the FRET community, the researchers are also working to establish suggestions for FRET microscopy.
Sanabria is planning to use his optical imaging technique on other biomolecules.
“This optical method can be used to study protein folding and misfolding or any structural organization of biomolecules,” added Sanabria. “It can also be used for drug screening and development, which requires knowing what a biomolecule looks like in order for a drug to target it.”
“This work is a milestone in structure determination using FRET to map short-lived functionally relevant enzyme states,” concluded Seidel.
Apart from Sanabria and Seidel, the study’s co-authors are Dmitro Rodnin, Katherina Hemmen, Thomas Peulen, Suren Felekyan, Mykola Dimura, and Ralf Kuhnemuth from the Institute of Molecular Physical Chemistry at Heinrich Heine University; Felix Koberling from PicoQuant GmbH; and Mark Fleissner and Wayne Hubbell from the Jules Stein Eye Institute at UCLA.
The study also included Holger Gohlke from the Institute of Pharmaceutical and Medicinal Chemistry at Heinrich Heine University.
The study was financed by the European Research Council via the Advanced Grant 2014 hybrid FRET (grant 671208 to CAMS) and the National Science Foundation CAREER MCB award for young investigators (grant 1749778 to HS).