Mar 3 2020
At times, biology can be fuzzy and, likewise, medicine involves highly complex mixtures of molecules that have to be dealt with.
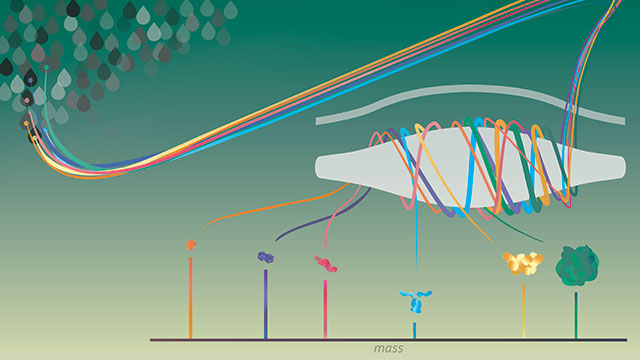 Single molecules of proteins or virus-like particles rotating around the commercially available Orbitrap analyzer to determine their exact mass. Image Credit: Northwestern University.
Single molecules of proteins or virus-like particles rotating around the commercially available Orbitrap analyzer to determine their exact mass. Image Credit: Northwestern University.
Northwestern University has developed a new technology that provides accurate measurements of proteins down to their atoms. This brings some level of clarity to researchers.
The new, robust technique, known as individual ion mass spectrometry, or I2MS for short, can establish the accurate mass of a wide range of intact proteins. This method weighs all molecules on an individual basis. This can help in interpreting infection and disease and can speed up vaccine design for coronavirus and other similar deadly viruses.
The I2MS technique offers a fundamentally new method to determine the weight of single molecules of proteins or the assemblies of proteins, such as whole viruses. Details of this method were recently published in the Nature Methods journal on March 2nd, 2020.
The method employs the Orbitrap mass analyzer system that is available in the market. The scientists demonstrated that their technique can be utilized on highly complex mixtures of intact proteins, and also on whole, virus particles that carry an assortment of cargo inside them.
This versatility and power will bring a new wave of molecular precision to a wide range of issues faced in disease biology, neurodegenerative plaques, virology, and vaccinology in general.
Quickly characterizing the masses of viruses and their infectious cargo over time may help scientists understand mutations that are occurring. Whether directly characterizing different strains of viruses or profiling different vaccine formulations, our new technology now can be deployed directly on these protein-containing samples to pursue the most urgent challenges of the day.
Neil L. Kelleher, Study Lead, Northwestern University
Kelleher established the top-down proteomics with one of the top teams across the world that focuses on intact proteins. Kelleher is also the Walter and Mary Elizabeth Glass Chair in Life Sciences in the departments of chemistry and molecular biosciences of the Weinberg College of Arts and Sciences.
The new technology will allow researchers to further interpret the composition of a virus’ exterior, known as the capsid, and also understand the infectious cargo held inside the capsid, added Kelleher. Because the scientists can examine a few single virus particles at a given time, they can obtain data about accurate alterations in every particle.
Many research groups are studying the use of viral capsids filled with cargo as a means to deliver life-saving drugs to patients. Our technology provides a practical way to determine if the cargo contains the correct drug or to find out what is actually within each virus particle.
Jared O. Kafader, Study First Author, Northwestern University
Kafader is also a senior research associate at Northwestern University’s Proteomics Center of Excellence. (Proteomics refers to the study and analysis of the structure and function of proteins.)
The Kelleher group performed a similar study that extends the analysis of I2MS to the fragmentation of intact species. This study was recently published in the Journal of Proteome Research.
Fragmentation of intact proteins can provide crucial information about where mutations or modifications occur on the protein and how they can be detected, stated the scientists. Such modifications could offer a better understanding of how proteins mutate or change in cancer patients.
The National Institute of General Medical Sciences, National Institutes of Health (grant P41 GM108569), the Sherman Fairchild Foundation, and Thermo Fisher Scientific supported the study.
The study is titled “Multiplexed mass spectrometry of individual ions improves measurement of proteoforms and their complexes.” Kelleher is the study’s corresponding author, and Kafader is the study’s first author.
Kelleher is also a professor in the department of medicine in the Feinberg School of Medicine. He is a member of the Robert H. Lurie Comprehensive Cancer Center of Northwestern University and the Chemistry of Life Processes Institute.